Note
Go to the end to download the full example code.
Plot events#
import numpy as np
import seaborn as sns
from bokeh.io import output_notebook
from bokeh.plotting import show
from systole.detection import ecg_peaks
from systole.plots import plot_events, plot_rr
from systole import import_dataset1
# Author: Nicolas Legrand <nicolas.legrand@cas.au.dk>
# Licence: GPL v3
Plot events distributions using Matplotlib as plotting backend#
ecg_df = import_dataset1(modalities=["ECG", "Stim"])
# Get events triggers
triggers_idx = [
np.where(ecg_df.stim.to_numpy() == 2)[0],
np.where(ecg_df.stim.to_numpy() == 1)[0],
]
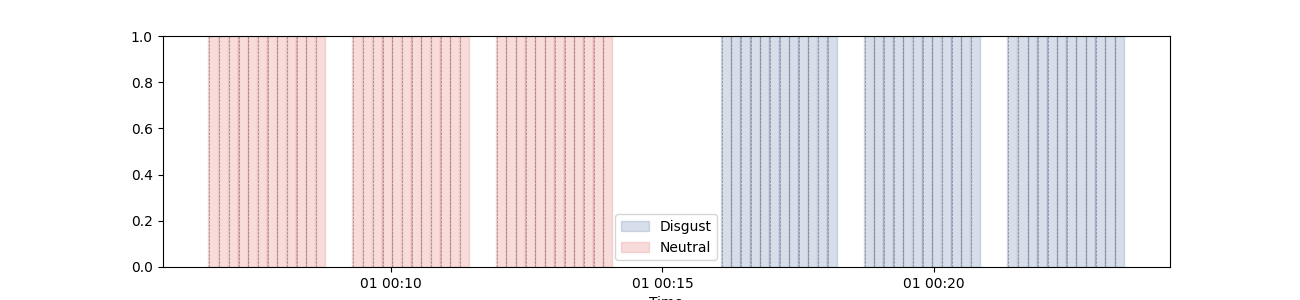
plot_events(
triggers_idx=triggers_idx,
labels=["Disgust", "Neutral"],
tmin=-0.5,
tmax=10.0,
figsize=(13, 3),
palette=[sns.xkcd_rgb["denim blue"], sns.xkcd_rgb["pale red"]],
)
0%| | 0/2 [00:00<?, ?it/s]
Downloading ECG channel: 0%| | 0/2 [00:00<?, ?it/s]
Downloading Stim channel: 0%| | 0/2 [00:00<?, ?it/s]
Downloading Stim channel: 100%|██████████| 2/2 [00:00<00:00, 14.72it/s]
Downloading Stim channel: 100%|██████████| 2/2 [00:00<00:00, 14.69it/s]
<Axes: xlabel='Time'>
Plot events distributions using Bokeh as plotting backend and add the RR time series#
output_notebook()
# Peak detection in the ECG signal using the Pan-Tompkins method
signal, peaks = ecg_peaks(ecg_df.ecg, method="pan-tompkins", sfreq=1000)
# First, we create a RR interval plot
rr_plot = plot_rr(peaks, input_type="peaks", backend="bokeh", figsize=250)
show(
# Then we add events annotations to this plot using the plot_events function
plot_events(
triggers_idx=triggers_idx,
backend="bokeh",
labels=["Disgust", "Neutral"],
tmin=-0.5,
tmax=10.0,
palette=[sns.xkcd_rgb["denim blue"], sns.xkcd_rgb["pale red"]],
ax=rr_plot.children[0],
)
)
Total running time of the script: (0 minutes 1.045 seconds)